Centromeres
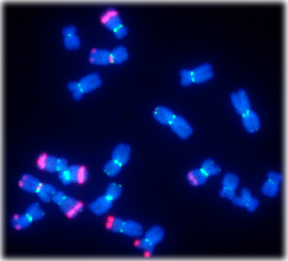
FISH image showing maize chromosomes. Centromeres (CentC) are green, knobs are red.
While kinetochore proteins are reasonably well conserved, centromeric DNAs are among the most poorly conserved sequences known; the homology is so slim that it is nearly impossible to identify centromeres based on sequence alone. Our strategy for finding centromeres is to start with the kinetochores and work down, i.e., to use anti-kinetochore antisera as a way to pull down the associated centromeric DNA.
The two main components of maize centromeres are a 156 bp tandem repeat known as CentC, and a centromere-specific retroelement known as CRM (Centromeric Retroelement in Maize). By immunoprecipitating nucleosomes (by ChIP) containing CENH3, we were able to confirm that CentC and CRM are indeed major centromeric repeats. These results have now been amply confirmed by the complete sequencing of two centromeres and ChIP-seq to precisely localize kinetochores over physical maps. Although CentC and CRM are the most conserved elements of maize centromeres, they are not required to recruit kinetochore proteins. Rather, in both plants and animals, centromeres are determined in a largely sequence-independent, or epigenetic fashion.
We have shown that RNA is probably one of the features that serves as an epigenetic mark for centromeres. By combining chromatin immunoprecipitation (ChIP) with RNA detection methods, we demonstrated that CRM as well as CentC are not only transcribed, but that much of the RNA remains tightly bound to the centromere/kinetochore complex. Very similar results were later obtained from human cells. We have recently shown that DNA binding of the key kinetochore protein CENPC is is strongly promoted by single stranded RNA, suggesting that RNA serves to reinforce the recruitment of inner kinetochore proteins. We are now proceeding to thoroughly investigate the structure and origin of centromeric RNAs.
 The Dawe Lab
The Dawe Lab