Review: Plant Mitosis
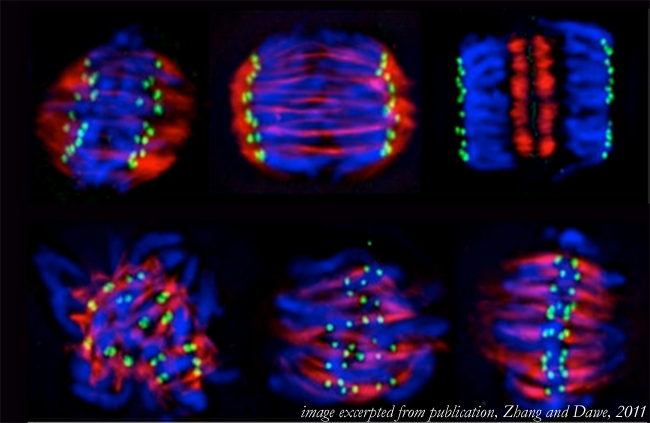
Mechanisms of plant spindle formation
Han Zhang and R. Kelly Dawe
From the issue entitled “Special issue on Mitotic Chromosomes and Microtubules: The Dynamic Duo”
Abstract
In eukaryotes, the formation of a bipolar spindle is necessary for the equal segregation of chromosomes to daughter cells. Chromosomes, microtubules and kinetochores all contribute to spindle morphogenesis and have important roles during mitosis. A unique property of flowering plant cells is that they entirely lack centrosomes, which in animals have a major role in spindle formation. The absence of these important structures suggests that plants have evolved novel mechanisms to assure chromosome segregation. In this review, we highlight some of the recent studies on plant mitosis and argue that plants utilize a variation of “spindle self-organization” that takes advantage of the early polarity of plant cells and accentuates the role of kinetochores in stabilizing the spindle midzone in prometaphase.
New Publication
Distinct influences of tandem repeats and retrotransposons on CENH3 nucleosome positioning.
Gent JI, Schneider KL, Topp CN, Rodriguez C, Presting GG, Dawe RK.
ABSTRACT:
BACKGROUND: Unique structural characteristics of centromere chromatin enable it to support assembly of the kinetochore and its associated tensions. The histone H3 variant CENH3 (centromeric histone H3) is viewed as the key element of centromere chromatin and its interaction with centromere DNA is epigenetic in that its localization to centromeres is not sequence-dependent.
RESULTS: In order to investigate what influence the DNA sequence exerts on CENH3 chromatin structure, we examined CENH3 nucleosome footprints on maize centromere DNA. We found a predominant average nucleosome spacing pattern of roughly 190-bp intervals, which was also the dominant arrangement for nucleosomes genome-wide. For CENH3-containing nucleosomes, distinct modes of nucleosome positioning were evident within that general spacing constraint. Over arrays of the major ~156-bp centromeric satellite sequence (tandem repeat) CentC, nucleosomes were not positioned in register with CentC monomers but in conformity with a striking ~10-bp periodicity of AA/TT dimers within the sequence. In contrast, nucleosomes on a class of centromeric retrotransposon (CRM2) lacked a detectable AA/TT periodicity but exhibited tightly phased positioning.
CONCLUSIONS: These data support a model in which general chromatin factors independent of both DNA sequence and CENH3 enforce roughly uniform centromeric nucleosome spacing while allowing flexibility in the mode in which nucleosomes are positioned. In the case of tandem repeat DNA, the natural bending effects related to AA/TT periodicity produce an energetically-favourable arrangement consistent with conformationally rigid nucleosomes and stable chromatin at centromeres.
 The Dawe Lab
The Dawe Lab